

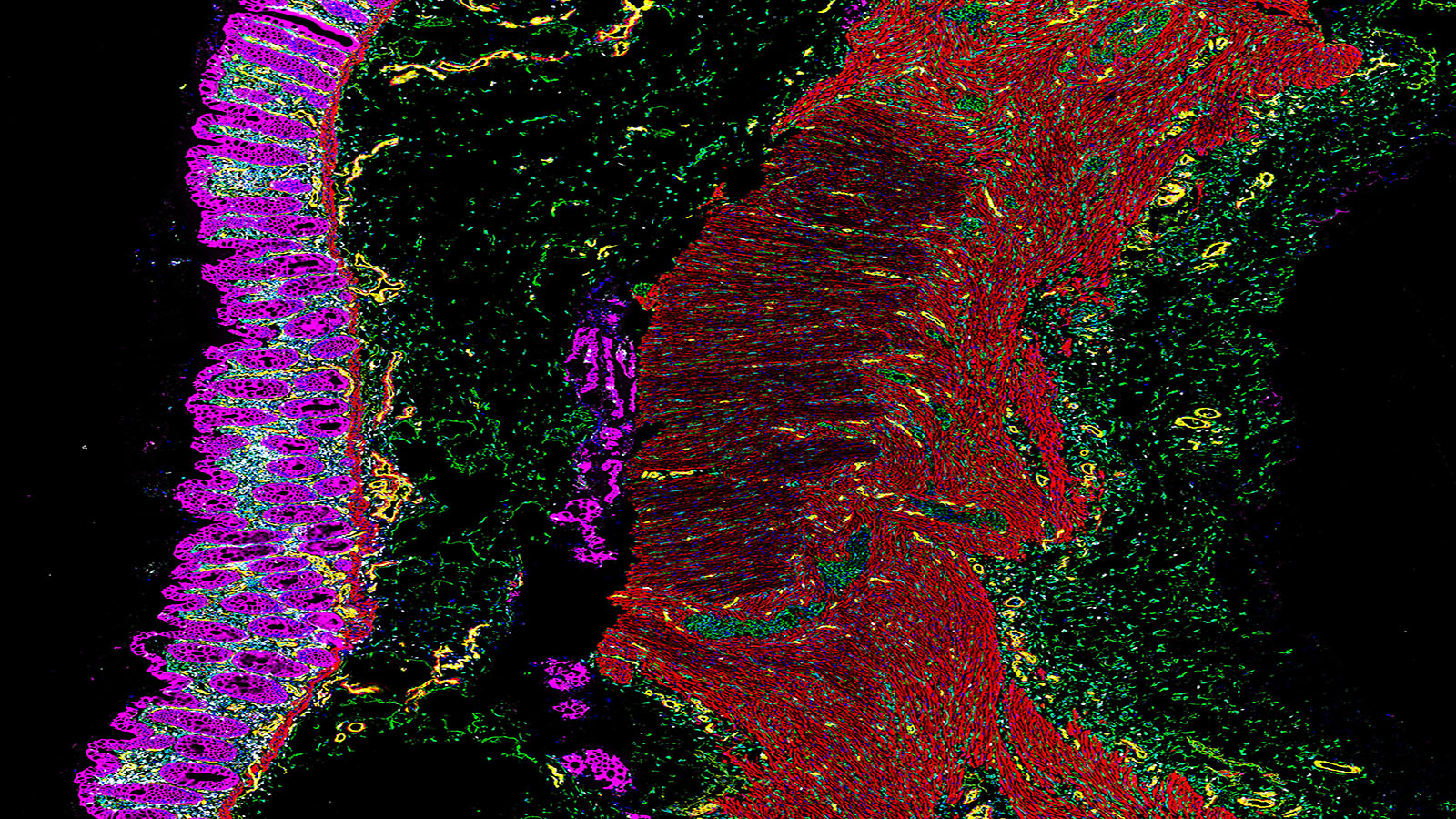
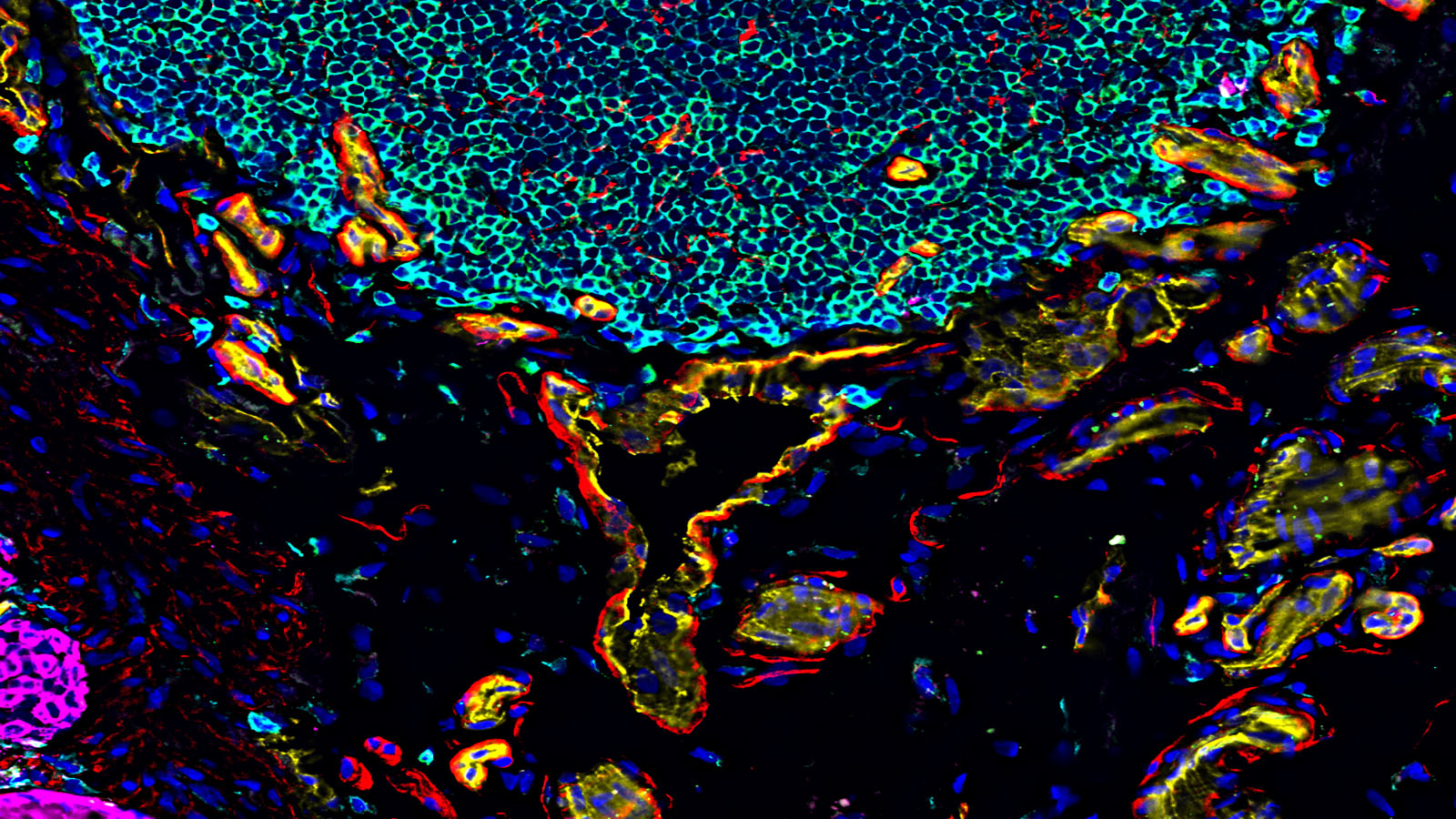
State of the art spatial biology is available with the Akoya Biosciences CODEX system. CODEX provides full spatial context and quantitative measurement of 40+ analytes from a single tissue slide. Single cell resolution is easily achieved. Application to clinical or discovery science is practical with validated performance on fresh frozen or FFPE tissue. Antibodies are available for mouse or human tissues. Furthermore, the assays are non-destructive, so tissue can be used for other downstream analysis or staining. More information is available at the Akoya Biosciences website.
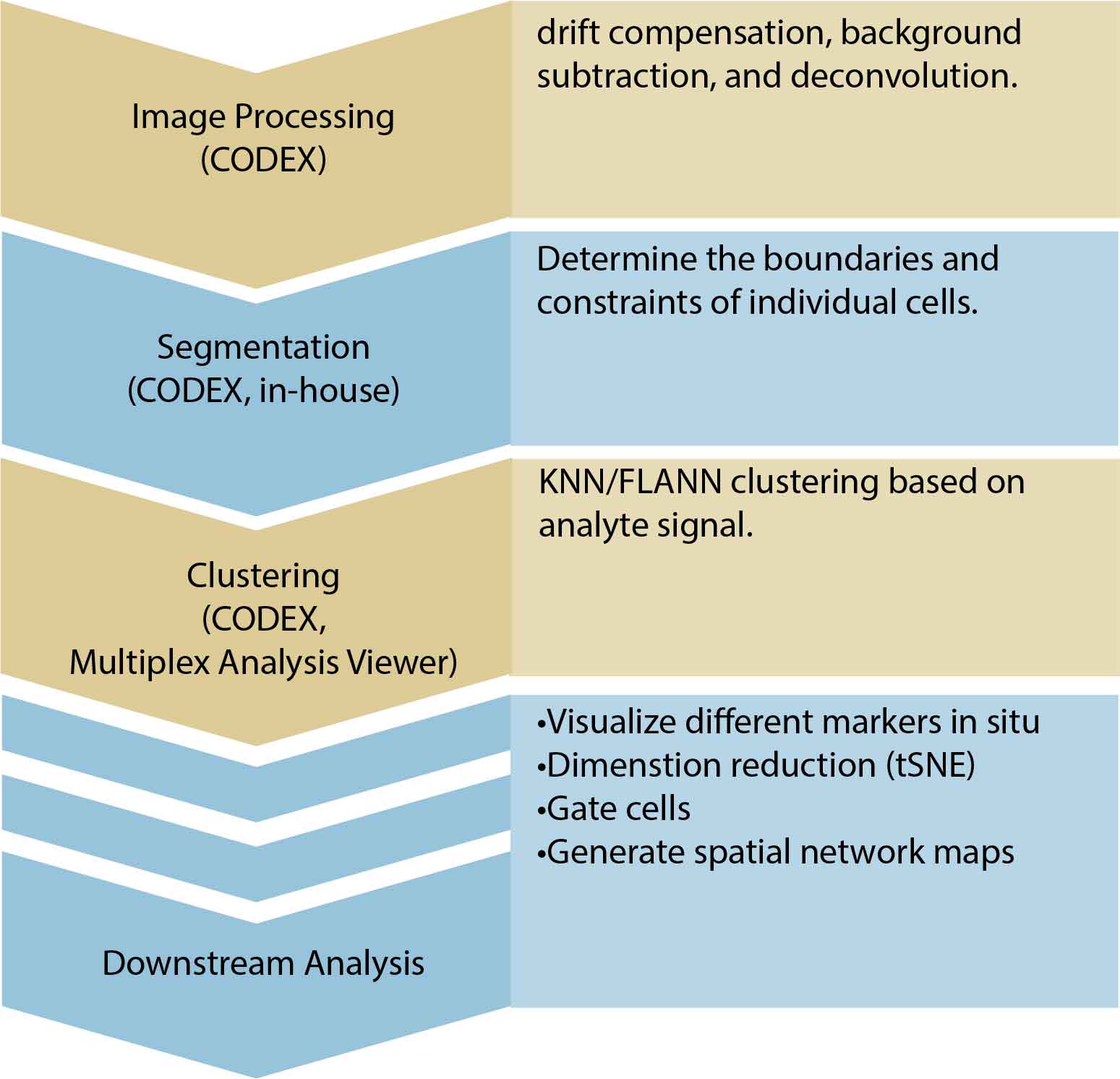
Raw CODEX images are processed and normalized in bulk. This includes drift compensation, background subtraction, and deconvolution. Cells are segmented based on either DAPI staining or membrane marker staining, and each of the analytes is quantified. A spatially annotated cell-by-analyte csv file will be returned. Processed image files are also returned for downstream analysis with any image software.

Revolutionary spatial biology awaits you with the NanoString GeoMx Digital Spatial Profiler. Given fresh frozen or FFPE tissue, the unique technology of GeoMx allows simultaneous, quantitative measurement of RNA and protein targets. The morphological context of the tissue can be resolved with customizable histology. Unlike CODEX, the GeoMx assay is conducted by region of interest (ROI). There are several strategies for choosing the ROIs to maximize return on the experiment design. Choose between curated panels of molecular targets or customize your own. Instructions for panel selection are available here. These data can also be integrated with Illumina high-throughput sequencing.
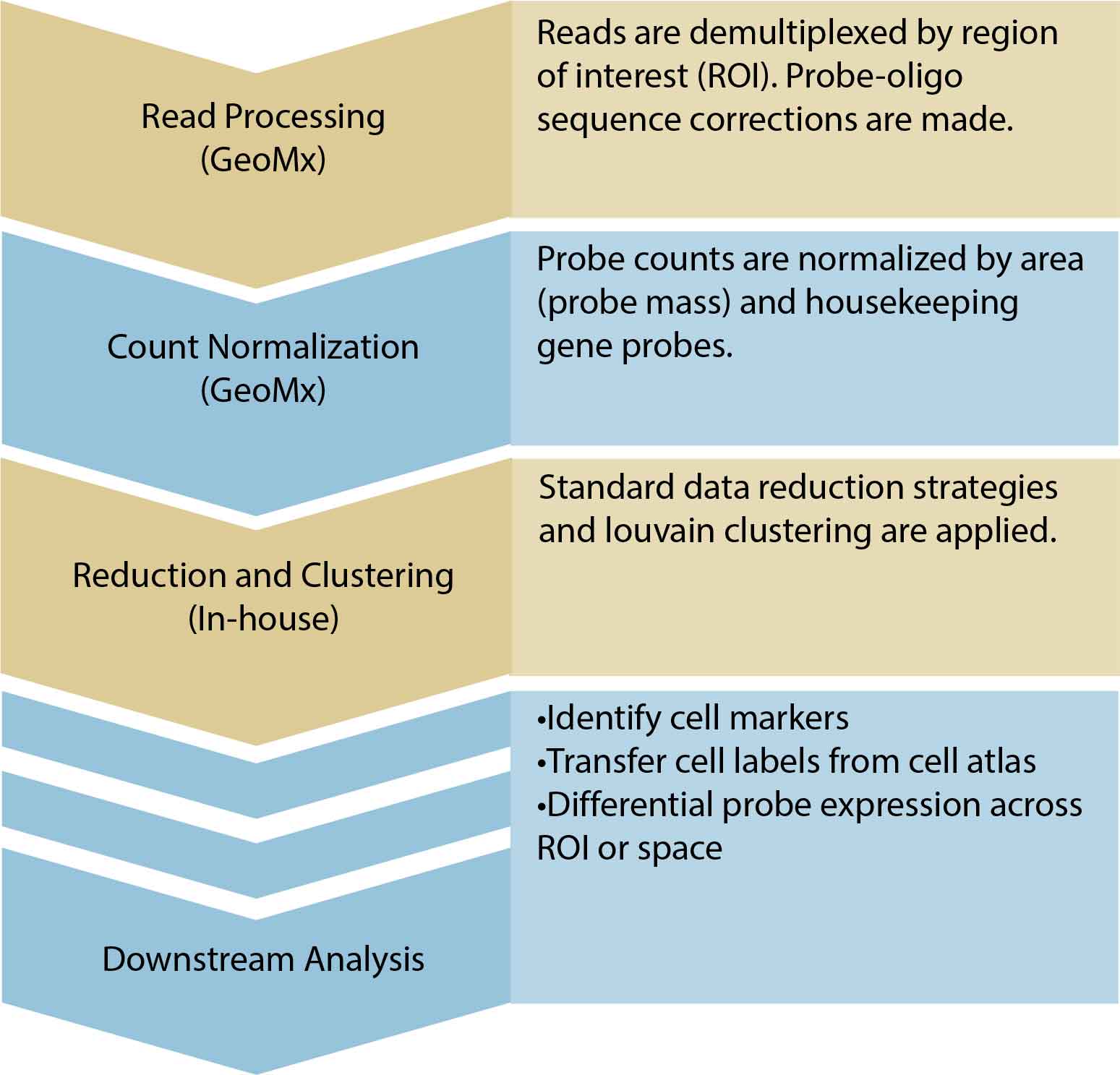
Raw morphology-stained images and demultiplexed probe count data will be made available. Corresponding spatial annotations will also be formatted. We will perform data quality control. The standard analysis workflow includes area scaling and control probe normalization. With the versatility of the GeoMx system, further downstream analysis will depend on ROI selection and experiment design.
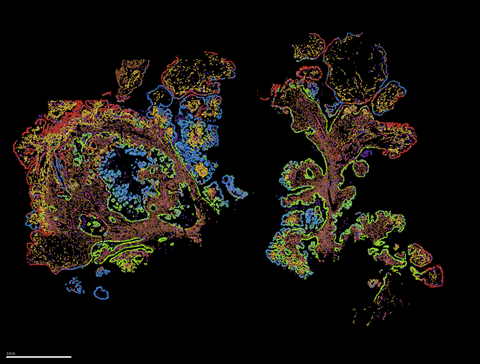
Discover the latest in single-cell spatial genomics with the MERSCOPE Platform. As the industry’s first high-plex in situ single-cell solution, it uses MERFISH technology to directly map and quantify the spatial distribution of RNA species in individual cells with subcellular resolution and without the need for downstream sequencing. The complete end-to-end solution includes the MERSCOPE Instrument, intuitive software, reagents, and consumables, making it easy to obtain and analyze high quality MERFISH measurements. With its versatility, high sensitivity and multiplexing power, and ease of use, the MERSCOPE is the ideal tool for your single-cell research needs. Let our Single Cell research core handle your MERSCOPE experiments for you. Learn more on the VizGen website here.
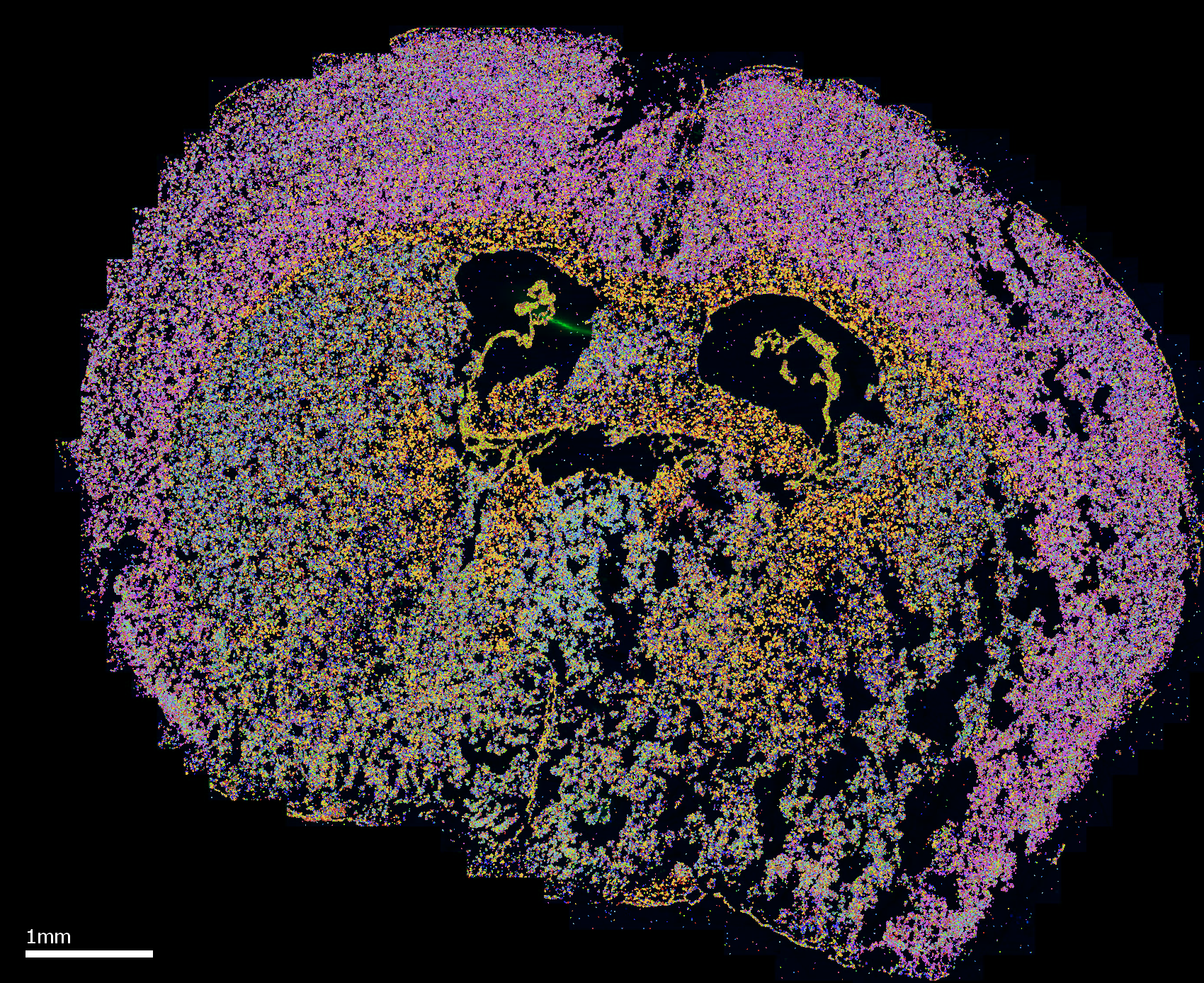
Unlock the full potential of your MERFISH data with the MERSCOPE™ Vizualizer software. This intuitive platform provides a powerful and user-friendly way to analyze and interpret the results of your MERSCOPE run. Explore the results of your experiment, from the detection of transcripts at the single-cell level to the visualization of cell clusters, with a suite of features that makes exploring your data easy and efficient. With its high-resolution images, heat maps, and scatterplots, the MERSCOPE™ Vizualizer software is the ideal tool for gaining valuable biological insights from your MERFISH experiment.
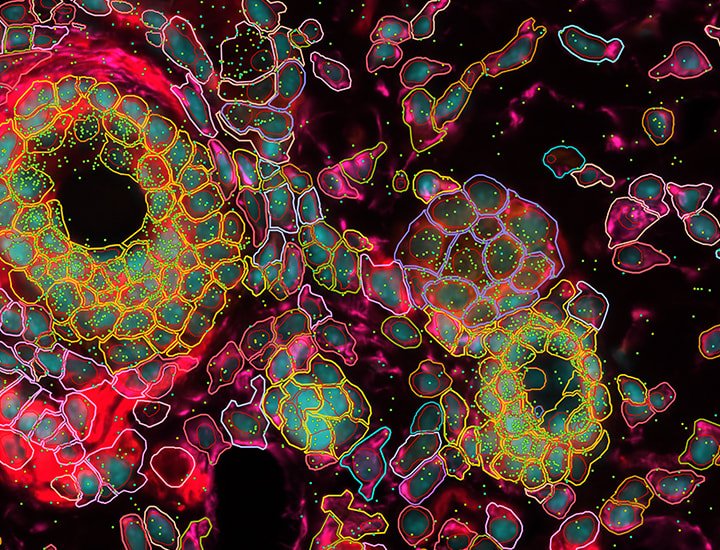
10x Genomics Xenium Analyzer provides easy to use, high throughput in situ profiling of hundreds of RNA targets at a subcellular resolution.
Supported assays:
Human or mouse pre-designed panels
Standalone or add-on custom panels of up to 500 genes
Xenium Prime 5K, with optional add-on custom panels of up to 100 genes
Sample Requirements
Compatible samples: FFPE or Fresh Frozen tissue
Mounted on Xenium slides
Sample area: 10.45mm x 22.45mm
Section thickness: FFPE 5 μm; Frozen 10 μm
Prior to sectioning onto Xenium slides, tissue morphology should be checked for any undesired artifacts
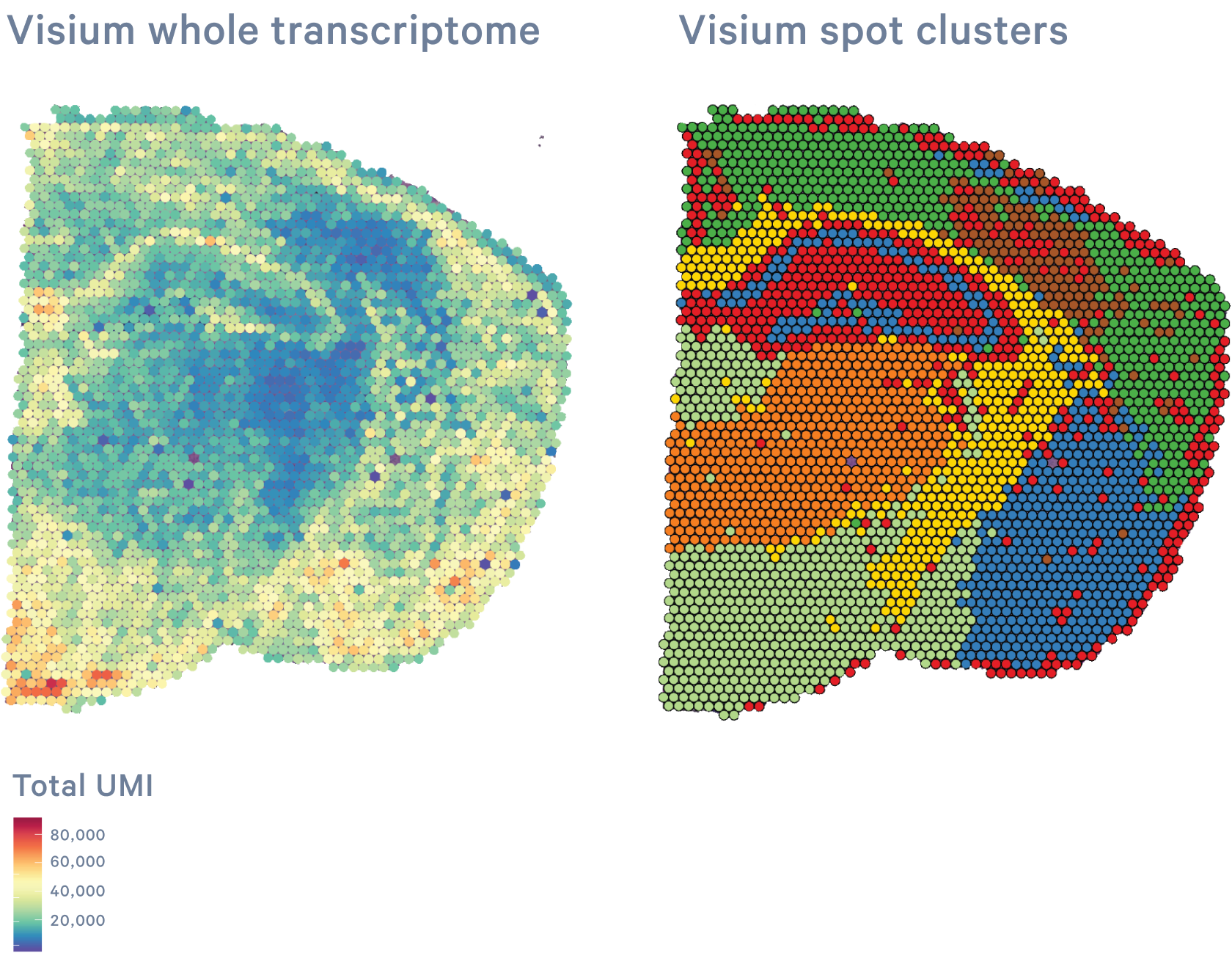
10x Genomics Visium CytAssist allows investigators to map the entire transcriptome of H&E- or immunofluorescently stained tissue sections with morphological context. The innovative CytoAssist technology expands sample accessibility, enabling the transfer of transcriptomic analytes from tissue sections on standard glass slides into the Visium workflow.
Supported assays:
Human or mouse whole transcriptome gene expression
Human or mouse whole transcriptome gene expression and targeted protein expression (not available for HD)
Sample Requirements
Compatible samples: FFPE, Fresh or Fixed Frozen tissue
Please ensure the desired tissue area will fit within the 6.5 x 6.5 mm or 11 x 11 mm capture area; only 6.5 x 6.5 mm is available for HD
There are two capture areas per slide
Tissue sections should be mounted within the allowable target area on the plain glass slide according to the 10x Genomics Tissue Preparation Guides
Recommended section thickness:
FFPE: 3-10 µm
FF or FxF: 10-20 µm
RNA quality of the block should be assessed prior to sectioning:
FFPE: DV200>30%
FF: RIN>4
FxF: DV200>50%